Note: Please see the update to this blog!
The best part about writing a dissertation is finding clever ways to procrastinate. The motivation for this blog comes from one of the more creative ways I’ve found to keep myself from writing. I’ve posted about data mining in the past and this post follows up on those ideas using a topic that is relevant to anyone that has ever considered getting, or has successfully completed, their PhD.
I think a major deterrent that keeps people away from graduate school is the requirement to write a dissertation or thesis. One often hears horror stories of the excessive page lengths that are expected. However, most don’t realize that dissertations are filled with lots of white space, e.g., pages are one-sided, lines are double-spaced, and the author can put any material they want in appendices. The actual written portion may only account for less than 50% of the page length. A single chapter may be 30-40 pages in length, whereas the same chapter published in the primary literature may only be 10 or so pages long in a journal. Regardless, students (myself included) tend to fixate on the ‘appropriate’ page length for a dissertation, as if it’s some sort of measure of how much work you’ve done to get your degree. Any professor will tell you that page length is not a good indicator of the quality of your work. Regardless, I feel that some general page length goal should be established prior to writing. This length could be a minimum to ensure you put forth enough effort, or an upper limit to ensure you aren’t too excessive on extraneous details.
It’s debatable as to what, if anything, page length indicates about the quality of one’s work. One could argue that it indicates absolutely nothing. My advisor once told me about a student in Chemistry that produced a dissertation that was less than five pages, and included nothing more than a molecular equation that illustrated the primary findings of the research. I’ve heard of other advisors that strongly discourage students from creating lengthy dissertations. Like any indicator, page length provides information that may or may not be useful. However, I guarantee that almost every graduate student has thought about an appropriate page length on at least one occasion during their education.
The University of Minnesota library system has been maintaining electronic dissertations since 2007 in their Digital Conservancy website. These digital archives represent an excellent opportunity for data mining. I’ve developed a data scraper that gathers information on student dissertations, such as page length, year and month of graduation, major, and primary advisor. Unfortunately, the code will not work unless you are signed in to the University of Minnesota library system. I’ll try my best to explain what the code does so others can use it to gather data on their own. I’ll also provide some figures showing some relevant data about dissertations. Obviously, this sample is not representative of all institutions or time periods, so extrapolation may be unwise. I also won’t be providing any of the raw data, since it isn’t meant to be accessible for those outside of the University system.
I’ll first show the code to get the raw data for each author. The code returns a list with two elements for each author. The first element has the permanent and unique URL for each author’s data and the second element contains a character string with relevant data to be parsed.
#import package
require(XML)
#starting URL to search
url.in<-'http://conservancy.umn.edu/handle/45273/browse-author?starts_with=0'
#output object
dat<-list()
#stopping criteria for search loop
stp.txt<-'2536-2536 of 2536.'
str.chk<-'foo'
#initiate search loop
while(!grepl(stp.txt,str.chk)){
html<-htmlTreeParse(url.in,useInternalNodes=T)
str.chk<-xpathSApply(html,'//p',xmlValue)[3]
names.tmp<-xpathSApply(html, "//table", xmlValue)[10]
names.tmp<-gsub("^\\s+", "",strsplit(names.tmp,'\n')[[1]])
names.tmp<-names.tmp[nchar(names.tmp)>0]
url.txt<-strsplit(names.tmp,', ')
url.txt<-lapply(
url.txt,
function(x){
cat(x,'\n')
flush.console()
#get permanent handle
url.tmp<-gsub(' ','+',x)
url.tmp<-paste(
'http://conservancy.umn.edu/handle/45273/items-by-author?author=',
paste(url.tmp,collapse='%2C+'),
sep=''
)
html.tmp<-readLines(url.tmp)
str.tmp<-rev(html.tmp[grep('handle',html.tmp)])[1]
str.tmp<-strsplit(str.tmp,'\"')[[1]]
str.tmp<-str.tmp[grep('handle',str.tmp)] #permanent URL
#parse permanent handle
perm.tmp<-htmlTreeParse(
paste('http://conservancy.umn.edu',str.tmp,sep=''),useInternalNodes=T
)
perm.tmp<-xpathSApply(perm.tmp, "//td", xmlValue)
perm.tmp<-perm.tmp[grep('Major|pages',perm.tmp)]
perm.tmp<-c(str.tmp,rev(perm.tmp)[1])
}
)
#append data to list, will contain some duplicates
dat<-c(dat,url.txt)
#reinitiate url search for next iteration
url.in<-strsplit(rev(names.tmp)[1],', ')[[1]]
url.in<-gsub(' ','+',url.in)
url.in<-paste(
'http://conservancy.umn.edu/handle/45273/browse-author?top=',
paste(url.in,collapse='%2C+'),
sep=''
)
}
#remove duplicates
dat<-unique(dat)
The basic approach is to use functions in the XML package to import and parse raw HTML from the web pages on the Digital Conservancy. This raw HTML is then further parsed using some of the base functions in R, such as grep and strsplit. The tricky part is to find the permanent URL for each student that contains the relevant information. I used the ‘browse by author’ search page as a starting point. Each ‘browse by author’ page contains links to 21 individuals. The code first imports the HTML, finds the permanent URL for each author, reads the HTML for each permanent URL, finds the relevant data for each dissertation, then continues with the next page of 21 authors. The loop stops once all records are imported.
The important part is to identify the format of each URL so the code knows where to look and where to re-initiate each search. For example, each author has a permanent URL that has the basic form http://conservancy.umn.edu/ plus ‘handle/12345’, where the last five digits are unique to each author (although the number of digits varied). Once the raw HTML is read in for each page of 21 authors, the code has to find text where the word ‘handle’ appears and then save the following digits to the output object. The permanent URL for each student is then accessed and parsed. The important piece of information for each student takes the following form:
University of Minnesota Ph.D. dissertation. July 2012. Major: Business. Advisor: Jane Doe. 1 computer file (PDF); iv, 147 pages, appendices A-B.
This code is found by searching the HTML for words like ‘Major’ or ‘pages’ after parsing the permanent URL by table cells (using the <td></td> tags). This chunk of text is then saved to the output object for additional parsing.
After the online data were obtained, the following code was used to identify page length, major, month of completion, year of completion, and advisor for each character string for each student. It looks messy but it’s designed to identify the data while handling as many exceptions as I was willing to incorporate into the parsing mechanism. It’s really nothing more than repeated calls to grep using appropriate search terms to subset the character string.
#function for parsing text from website
get.txt<-function(str.in){
#separate string by spaces
str.in<-strsplit(gsub(',',' ',str.in,fixed=T),' ')[[1]]
str.in<-gsub('.','',str.in,fixed=T)
#get page number
pages<-str.in[grep('page',str.in)[1]-1]
if(grepl('appendices|appendix|:',pages)) pages<-NA
#get major, exception for error
if(class(try({
major<-str.in[c(
grep(':|;',str.in)[1]:(grep(':|;',str.in)[2]-1)
)]
major<-gsub('.','',gsub('Major|Mayor|;|:','',major),fixed=T)
major<-paste(major[nchar(major)>0],collapse=' ')
}))=='try-error') major<-NA
#get year of graduation
yrs<-seq(2006,2013)
yr<-str.in[grep(paste(yrs,collapse='|'),str.in)[1]]
yr<-gsub('Major|:','',yr)
if(!length(yr)>0) yr<-NA
#get month of graduation
months<-c('January','February','March','April','May','June','July','August',
'September','October','November','December')
month<-str.in[grep(paste(months,collapse='|'),str.in)[1]]
month<-gsub('dissertation|dissertatation|\r\n|:','',month)
if(!length(month)>0) month<-NA
#get advisor, exception for error
if(class(try({
advis<-str.in[(grep('Advis',str.in)+1):(grep('computer',str.in)-2)]
advis<-paste(advis,collapse=' ')
}))=='try-error') advis<-NA
#output text
c(pages,major,yr,month,advis)
}
#get data using function, ran on 'dat'
check.pgs<-do.call('rbind',
lapply(dat,function(x){
cat(x[1],'\n')
flush.console()
c(x[1],get.txt(x[2]))})
)
#convert to dataframe
check.pgs<-as.data.frame(check.pgs,sringsAsFactors=F)
names(check.pgs)<-c('handle','pages','major','yr','month','advis')
#reformat some vectors for analysis
check.pgs$pages<-as.numeric(as.character(check.pgs$pages))
check.pgs<-na.omit(check.pgs)
months<-c('January','February','March','April','May','June','July','August',
'September','October','November','December')
check.pgs$month<-factor(check.pgs$month,months,months)
check.pgs$major<-tolower(check.pgs$major)
The section of the code that begins with #get data using function takes the online data (stored as dat on my machine) and applies the function to identify the relevant information. The resulting text is converted to a data frame and some minor reworkings are applied to convert some vectors to numeric or factor values. Now the data are analyzed using the check.pgs object.
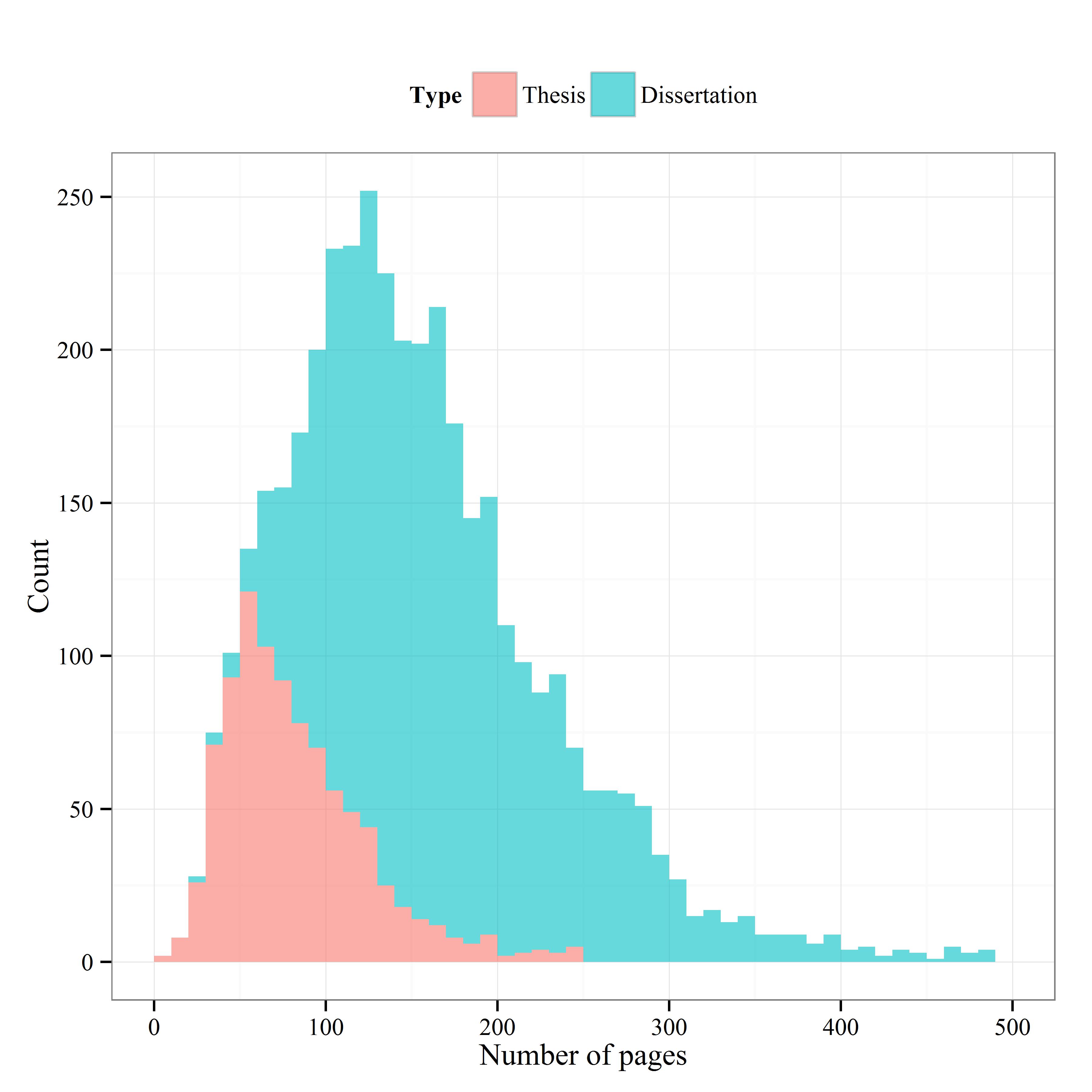
The data contained 2,536 records for students that completed their dissertations since 2007. The range was incredibly variable (minimum of 21 pages, maximum of 2002), but most dissertations were around 100 to 200 pages.

Interestingly, a lot of students graduated in August just prior to the fall semester. As expected, spikes in defense dates were also observed in December and May at the ends of the fall and spring semesters.

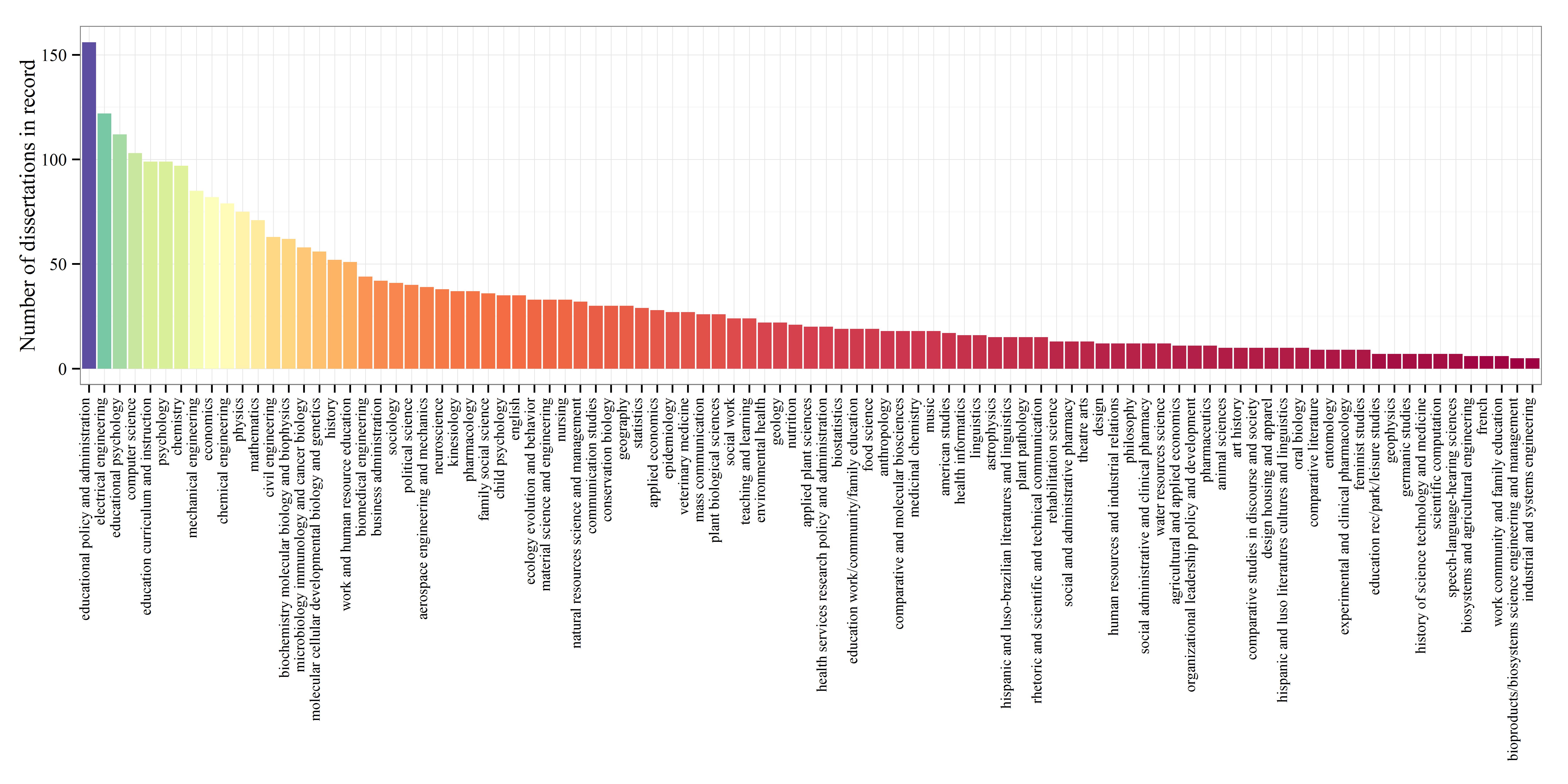
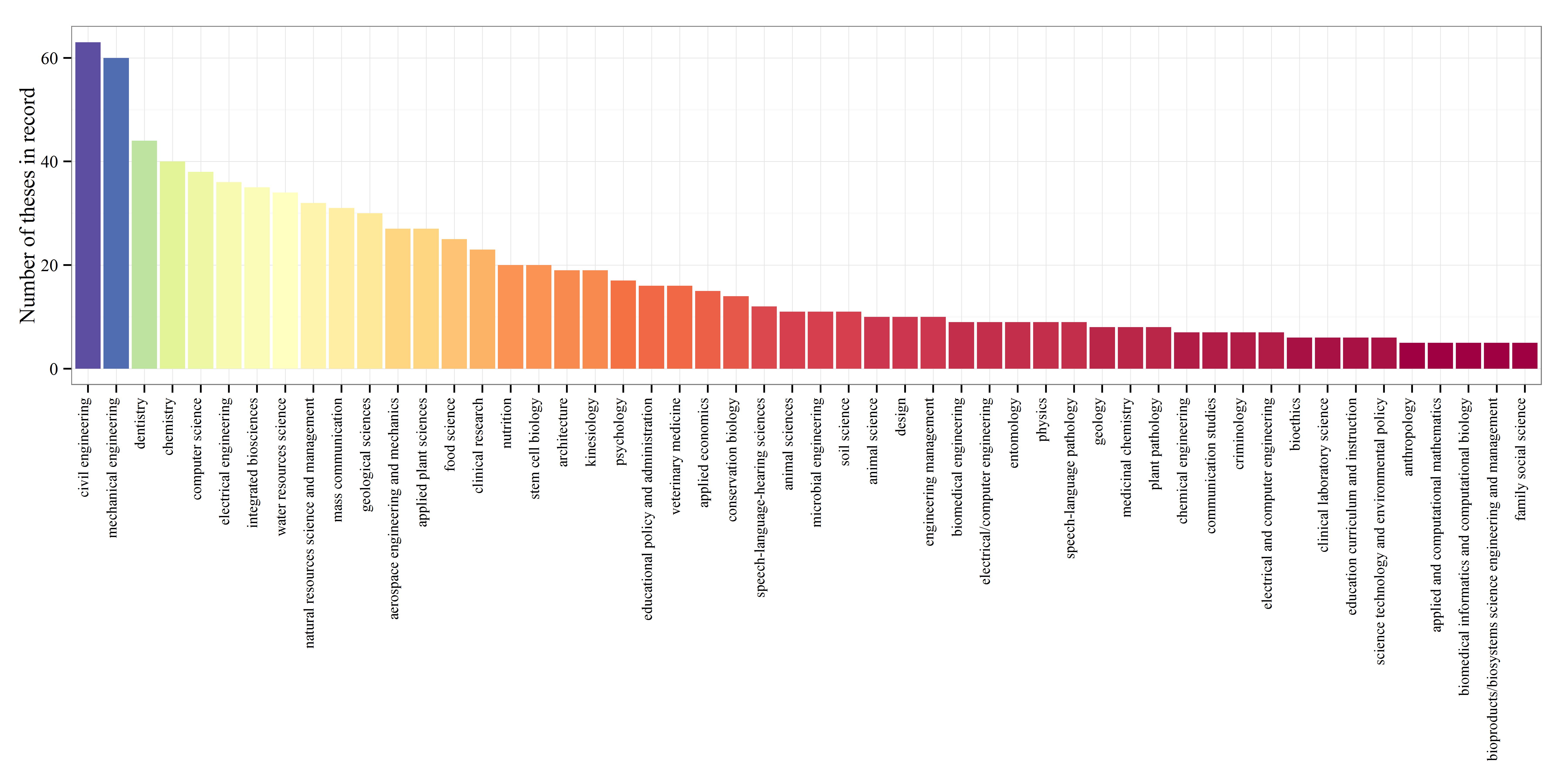
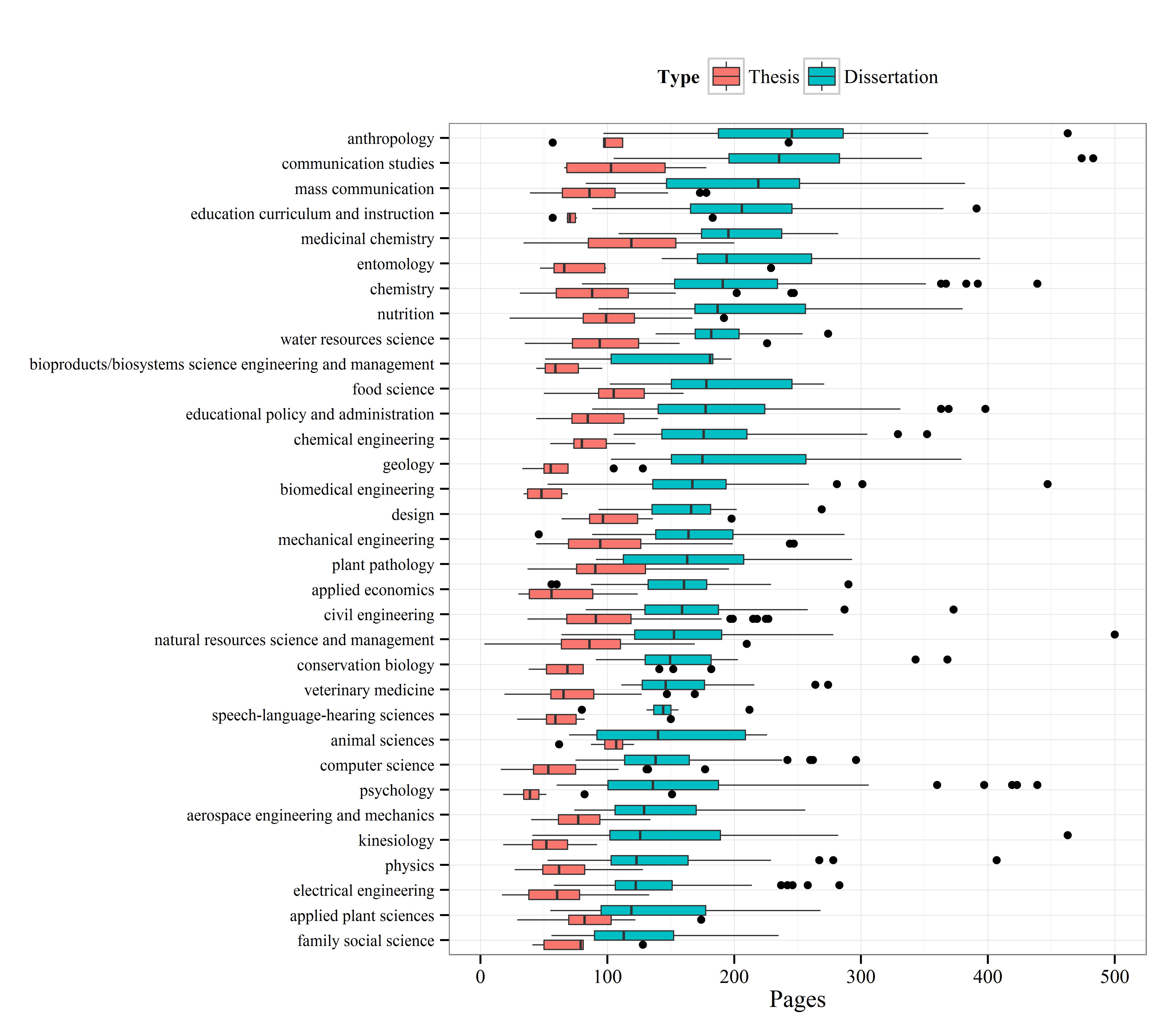
The top four majors with the most dissertations on record were (in descending order) educational policy and administration, electrical engineering, educational psychology, and psychology.

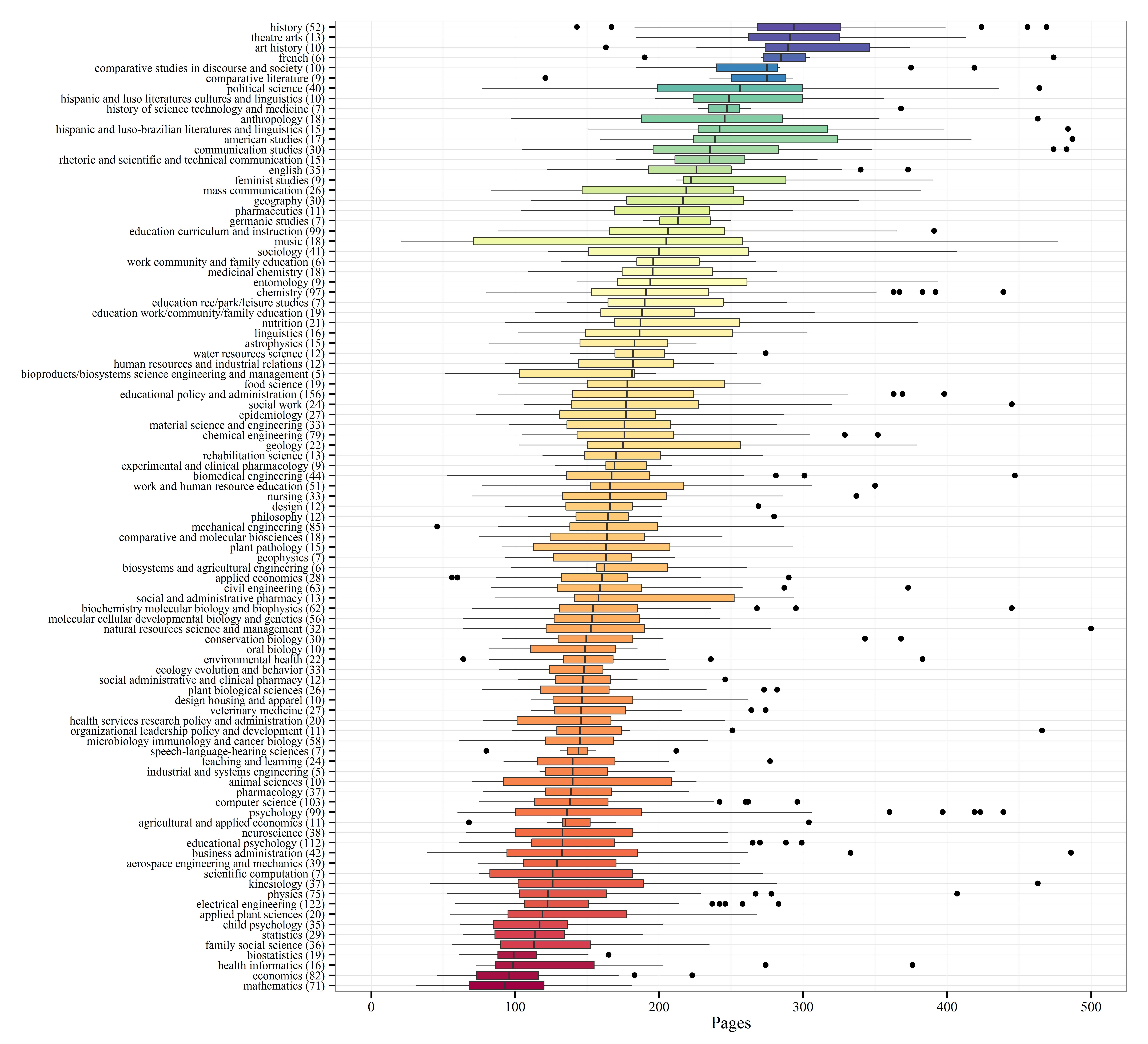
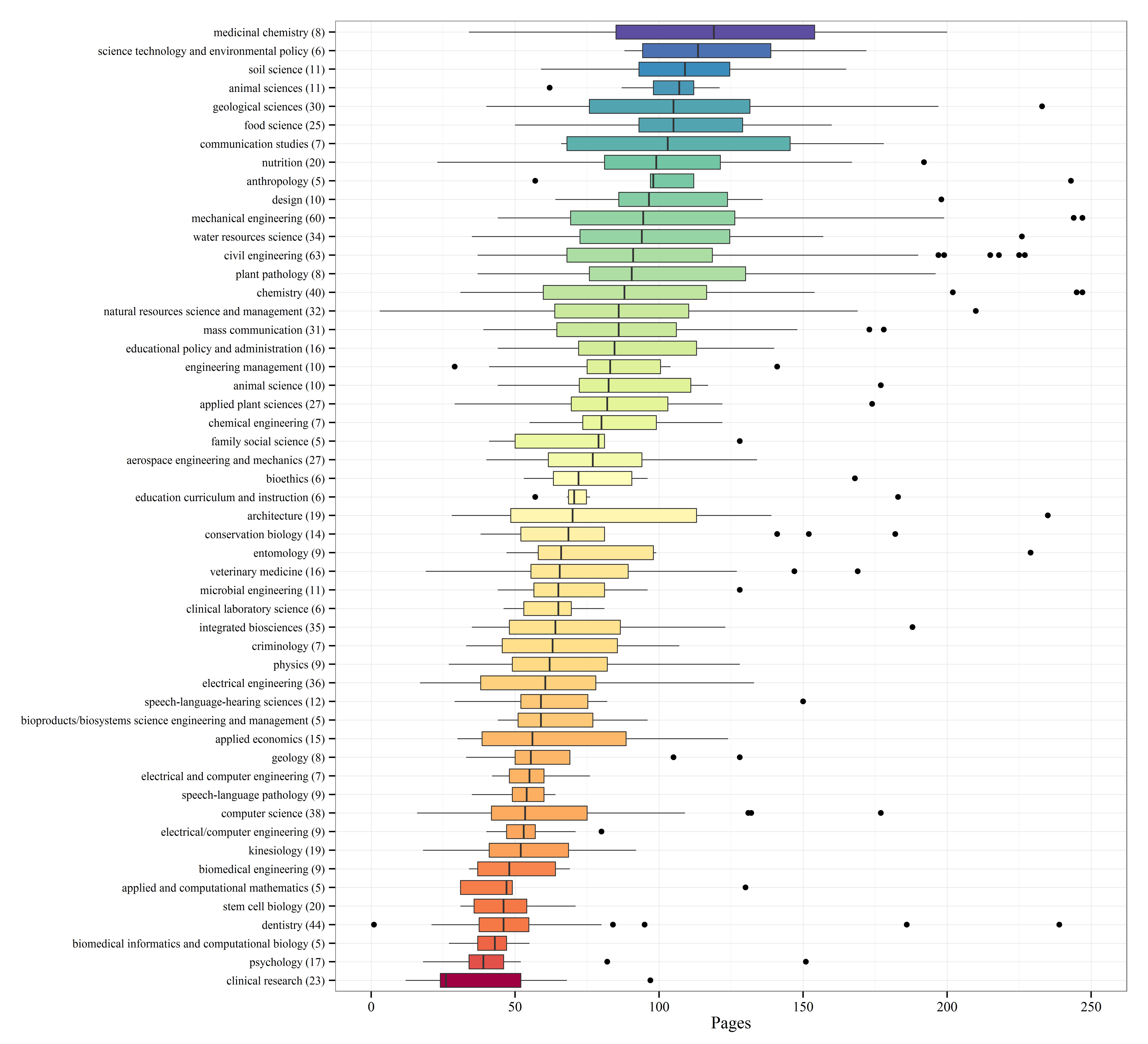
I’ve selected the top fifty majors with the highest number of dissertations and created boxplots to show relative distributions. Not many differences are observed among the majors, although some exceptions are apparent. Economics, mathematics, and biostatistics had the lowest median page lengths, whereas anthropology, history, and political science had the highest median page lengths. This distinction makes sense given the nature of the disciplines.

I’ve also completed a count of number of students per advisor. The maximum number of students that completed their dissertations for a single advisor since 2007 was eight. Anyhow, I’ve satiated my curiosity on this topic so it’s probably best that I actually work on my own dissertation rather than continue blogging. For those interested, the below code was used to create the plots.
######
#plot summary of data
require(ggplot2)
mean.val<-round(mean(check.pgs$pages))
med.val<-median(check.pgs$pages)
sd.val<-round(sd(check.pgs$pages))
rang.val<-range(check.pgs$pages)
txt.val<-paste('mean = ',mean.val,'\nmed = ',med.val,'\nsd = ',sd.val,
'\nmax = ',rang.val[2],'\nmin = ', rang.val[1],sep='')
#histogram for all
hist.dat<-ggplot(check.pgs,aes(x=pages))
pdf('C:/Users/Marcus/Desktop/hist_all.pdf',width=7,height=5)
hist.dat + geom_histogram(aes(fill=..count..),binwidth=10) +
scale_fill_gradient("Count", low = "blue", high = "green") +
xlim(0, 500) + geom_text(aes(x=400,y=100,label=txt.val))
dev.off()
#barplot by month
month.bar<-ggplot(check.pgs,aes(x=month,fill=..count..))
pdf('C:/Users/Marcus/Desktop/month_bar.pdf',width=10,height=5.5)
month.bar + geom_bar() + scale_fill_gradient("Count", low = "blue", high = "green")
dev.off()
######
#histogram by most popular majors
#sort by number of dissertations by major
get.grps<-list(c(1:4),c(5:8))#,c(9:12),c(13:16))
for(val in 1:length(get.grps)){
pop.maj<-names(sort(table(check.pgs$major),decreasing=T)[get.grps[[val]]])
pop.maj<-check.pgs[check.pgs$major %in% pop.maj,]
pop.med<-aggregate(pop.maj$pages,list(pop.maj$major),function(x) round(median(x)))
pop.n<-aggregate(pop.maj$pages,list(pop.maj$major),length)
hist.maj<-ggplot(pop.maj, aes(x=pages))
hist.maj<-hist.maj + geom_histogram(aes(fill = ..count..), binwidth=10)
hist.maj<-hist.maj + facet_wrap(~major,nrow=2,ncol=2) + xlim(0, 500) +
scale_fill_gradient("Count", low = "blue", high = "green")
y.txt<-mean(ggplot_build(hist.maj)$panel$ranges[[1]]$y.range)
txt.dat<-data.frame(
x=rep(450,4),
y=rep(y.txt,4),
major=pop.med$Group.1,
lab=paste('med =',pop.med$x,'\nn =',pop.n$x,sep=' ')
)
hist.maj<-hist.maj + geom_text(data=txt.dat, aes(x=x,y=y,label=lab))
out.name<-paste('C:/Users/Marcus/Desktop/group_hist',val,'.pdf',sep='')
pdf(out.name,width=9,height=7)
print(hist.maj)
dev.off()
}
######
#boxplots of data for fifty most popular majors
pop.maj<-names(sort(table(check.pgs$major),decreasing=T)[1:50])
pop.maj<-check.pgs[check.pgs$major %in% pop.maj,]
pdf('C:/Users/Marcus/Desktop/pop_box.pdf',width=11,height=9)
box.maj<-ggplot(pop.maj, aes(factor(major), pages, fill=pop.maj$major))
box.maj<-box.maj + geom_boxplot(lwd=0.5) + ylim(0,500) + coord_flip()
box.maj + theme(legend.position = "none", axis.title.y=element_blank())
dev.off()
Update: By popular request, I’ve redone the boxplot summary with major sorted by median page length.