Note: Please see the update to this post!
Neural networks have received a lot of attention for their abilities to ‘learn’ relationships among variables. They represent an innovative technique for model fitting that doesn’t rely on conventional assumptions necessary for standard models and they can also quite effectively handle multivariate response data. A neural network model is very similar to a non-linear regression model, with the exception that the former can handle an incredibly large amount of model parameters. For this reason, neural network models are said to have the ability to approximate any continuous function.
I’ve been dabbling with neural network models for my ‘research’ over the last few months. I’ll admit that I was drawn to the approach given the incredible amount of hype and statistical voodoo that is attributed to these models. After working with neural networks, I’ve found that they are often no better, and in some cases much worse, than standard statistical techniques for predicting relationships among variables. Regardless, the foundational theory of neural networks is pretty interesting, especially when you consider how computer science and informatics has improved our ability to create useful models.
R has a few packages for creating neural network models (neuralnet, nnet, RSNNS). I have worked extensively with the nnet package created by Brian Ripley. The functions in this package allow you to develop and validate the most common type of neural network model, i.e, the feed-forward multi-layer perceptron. The functions have enough flexibility to allow the user to develop the best or most optimal models by varying parameters during the training process. One major disadvantage is an inability to visualize the models. In fact, neural networks are commonly criticized as ‘black-boxes’ that offer little insight into causative relationships among variables. Recent research, primarily in the ecological literature, has addressed this criticism and several approaches have since been developed to ‘illuminate the black-box’.1
As far as I know, none of the recent techniques for evaluating neural network models are available in R. Özesmi and Özemi (1999)2 describe a neural interpretation diagram (NID) to visualize parameters in a trained neural network model. These diagrams allow the modeler to qualitatively examine the importance of explanatory variables given their relative influence on response variables, using model weights as inference into causation. More specifically, the diagrams illustrate connections between layers with width and color in proportion to magnitude and direction of each weight. More influential variables would have lots of thick connections between the layers.
In this blog I present a function for plotting neural networks from the nnet package. This function allows the user to plot the network as a neural interpretation diagram, with the option to plot without color-coding or shading of weights. The neuralnet package also offers a plot method for neural network objects and I encourage interested readers to check it out. I have created the function for the nnet package given my own preferences for aesthetics, so its up to the reader to choose which function to use. I plan on preparing this function as a contributed package to CRAN, but thought I’d present it in my blog first to gauge interest.
Let’s start by creating an artificial dataset to build the model. This is a similar approach that I used in my previous blog about collinearity. We create eight random variables with an arbitrary correlation struction and then create a response variable as a linear combination of the eight random variables.
require(clusterGeneration)
set.seed(2)
num.vars<-8
num.obs<-1000
#arbitrary correlation matrix and random variables
cov.mat<-genPositiveDefMat(num.vars,covMethod=c("unifcorrmat"))$Sigma
rand.vars<-mvrnorm(num.obs,rep(0,num.vars),Sigma=cov.mat)
parms<-runif(num.vars,-10,10)
#response variable as linear combination of random variables and random error term
y<-rand.vars %*% matrix(parms) + rnorm(num.obs,sd=20)
Now we can create a neural network model using our synthetic dataset. The nnet function can input a formula or two separate arguments for the response and explanatory variables (we use the latter here). We also have to convert the response variable to a 0-1 continuous scale in order to use the nnet function properly (via the linout argument, see the documentation).
require(nnet) rand.vars<-data.frame(rand.vars) y<-data.frame((y-min(y))/(max(y)-min(y))) names(y)<-'y' mod1<-nnet(rand.vars,y,size=10,linout=T)
We’ve created a neural network model with ten nodes in the hidden layer and a linear transfer function for the response variable. All other arguments are as default. The tricky part of developing an optimal neural network model is identifying a combination of parameters that produces model predictions closest to observed. Keeping all other arguments as default is not a wise choice but is a trivial matter for this blog. Next, we use the plot function now that we have a neural network object.
First we import the function from my Github account (aside: does anyone know a better way to do this?).
#import function from Github
require(RCurl)
root.url<-'https://gist.githubusercontent.com/fawda123'
raw.fun<-paste(
root.url,
'5086859/raw/cc1544804d5027d82b70e74b83b3941cd2184354/nnet_plot_fun.r',
sep='/'
)
script<-getURL(raw.fun, ssl.verifypeer = FALSE)
eval(parse(text = script))
rm('script','raw.fun')
The function is now loaded in our workspace as plot.nnet. We can use the function just by calling plot since it recognizes the neural network object as input.
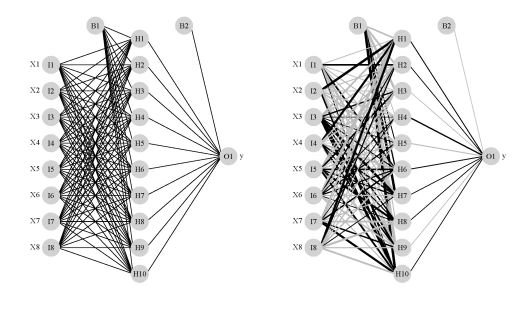
par(mar=numeric(4),mfrow=c(1,2),family='serif') plot(mod1,nid=F) plot(mod1)
The image on the left is a standard illustration of a neural network model and the image on the right is the same model illustrated as a neural interpretation diagram (default plot). The black lines are positive weights and the grey lines are negative weights. Line thickness is in proportion to magnitude of the weight relative to all others. Each of the eight random variables are shown in the first layer (labelled as X1-X8) and the response variable is shown in the far right layer (labelled as ‘y’). The hidden layer is labelled as H1 through H10, which was specified using the size argument in the nnet function. B1 and B2 are bias layers that apply constant values to the nodes, similar to intercept terms in a regression model.
The function has several arguments that affect the plotting method:
mod.in |
model object for input created from nnet function |
nid |
logical value indicating if neural interpretation diagram is plotted, default T |
all.out |
logical value indicating if all connections from each response variable are plotted, default T |
all.in |
logical value indicating if all connections to each input variable are plotted, default T |
wts.only |
logical value indicating if connections weights are returned rather than a plot, default F |
rel.rsc |
numeric value indicating maximum width of connection lines, default 5 |
circle.cex |
numeric value indicating size of nodes, passed to cex argument, default 5 |
node.labs |
logical value indicating if text labels are plotted, default T |
line.stag |
numeric value that specifies distance of connection weights from nodes |
cex.val |
numeric value indicating size of text labels, default 1 |
alpha.val |
numeric value (0-1) indicating transparency of connections, default 1 |
circle.col |
text value indicating color of nodes, default ‘lightgrey’ |
pos.col |
text value indicating color of positive connection weights, default ‘black’ |
neg.col |
text value indicating color of negative connection weights, default ‘grey’ |
... |
additional arguments passed to generic plot function |
Most of the arguments can be tweaked for aesthetics. We’ll illustrate using a neural network model created in the example code for the nnet function:
#example data and code from nnet function examples
ir<-rbind(iris3[,,1],iris3[,,2],iris3[,,3])
targets<-class.ind( c(rep("s", 50), rep("c", 50), rep("v", 50)) )
samp<-c(sample(1:50,25), sample(51:100,25), sample(101:150,25))
ir1<-nnet(ir[samp,], targets[samp,], size = 2, rang = 0.1,decay = 5e-4, maxit = 200)
#plot the model with different default values for the arguments
par(mar=numeric(4),family='serif')
plot.nnet(ir1,pos.col='darkgreen',neg.col='darkblue',alpha.val=0.7,rel.rsc=15,
circle.cex=10,cex=1.4,
circle.col='brown')
The neural network plotted above shows how we can tweak the arguments based on our preferences. This figure also shows that the function can plot neural networks with multiple response variables (‘c’, ‘s’, and ‘v’ in the iris dataset).
Another useful feature of the function is the ability to get the connection weights from the original nnet object. Admittedly, the weights are an attribute of the original function but they are not nicely arranged. We can get the weight values directly with the plot.nnet function using the wts.only argument.
plot.nnet(ir1,wts.only=T) $`hidden 1` [1] 0.2440625 0.5161636 1.9179850 -2.8496175 [5] -1.4606777 $`hidden 2` [1] 9.222086 6.350143 7.896035 -11.666666 [5] -8.531172 $`out 1` [1] -5.868639 -10.334504 11.879805 $`out 2` [1] -4.6083813 8.8040909 0.1754799 $`out 3` [1] 6.2251557 -0.3604812 -12.7215625
The function returns a list with all connections to the hidden layer (hidden 1 through hidden 2) and all connections to the output layer (out1 through out3). The first value in each element of the list is the weight from the bias layer.
The last features of the plot.nnet function we’ll look at are the all.in and all.out arguments. We can use these arguments to plot connections for specific variables of interest. For example, what if we want to examine the weights that are relevant only for sepal width (‘Sepal W.’) and virginica sp. (‘v’)?
plot.nnet(ir1,pos.col='darkgreen',neg.col='darkblue',alpha.val=0.7,rel.rsc=15, circle.cex=10,cex=1.4,circle.col='brown',all.in='Sepal W.',all.out='v')
This exercise is relatively trivial for a small neural network model but can be quite useful for a larger model.
Feel free to grab the function from Github (linked above). I encourage suggestions on ways to improve its functionality. I will likely present more quantitative methods of evaluating neural networks in a future blog, so stay tuned!
1Olden, J.D., Jackson, D.A. 2002. Illuminating the ‘black-box’: a randomization approach for understanding variable contributions in artificial neural networks. Ecological Modelling. 154:135-150.
2Özesmi, S.L., Özesmi, U. 1999. An artificial neural network approach to spatial habitat modeling with interspecific interaction. Ecological Modelling. 116:15-31.



Fantastic blog post and code.
I used neuralnet simply so I could plot the network however my preference is for nnet package. I’m not sure why but nnet seems easier to train.
It’d be good to get this functionality in the main R package.
Thank you for this post, it’s very interesting
Few bugs :
You need to add
require(clusterGeneration) # for access to genPositiveDefMat function
I had this error “requires numeric matrix/vector arguments” for this line :
y<-rand.vars %*% matrix(parms) + rnorm(num.obs,sd=20)
Which i fixed this way, it didn't like the dataframe class for %*% :
y<-as.matrix(rand.vars) %*% matrix(parms) + rnorm(num.obs,sd=20)
And also an error on the plot
plot(nnet.mod1,nid=F)
Fixed by :
plot(mod1, nid = F)
Thanks Pierre, my mistake. I’ve made the changes, should work correctly now.
Visualizing neural networks from the nnet package | spider's space
“First we import the function from my Github account (aside: does anyone know a better way to do this?”
I’d try this:
require(devtools)
source_gist(“5086859”)
Cheers
harald
Excellent, thanks Harald. I’ll try that in future posts.
Thanks for your interesting post! I adopted this idea for the neuralnet package: https://gist.github.com/sjewo/5099683
if you have a very large dataset with like 500+ variables (predictors), can neural networks give me a list of variable importance and a metric on the optimal number of variables to use.
Like in randomForest package it gives you the variable importance and additional package AUCRF gives me a optimal set of variables
I have some code I might share in future posts about variable importance values for neural networks. The optimal number of variables is a more tricky issue… stay tuned.
I’ve found some strange error:
df <- data.frame(x=1:10, y=sqrt(1:10))
n <- nnet(y ~ x, data=df, size=2)
plot(n) # OK
n <- nnet(as.formula('y ~ x'), data=df, size=2)
plot(n) # Almost OK, broken input and output labels
f <- as.formula('y ~ x')
n <- nnet(f, data=df, size=2)
plot(n) # Error: object of type 'symbol' is not subsettable
Ah, thanks for pointing this out. I still need to tweak the function to deal with different methods for the nnet object.
Alright, I was missing a call to
evalon line 56 of the function on the Github account. This should work correctly now.Terrific blog post! This is really a fantastic tool that I’ve put to use already. Thanks.
Animating neural networks from the nnet package – R is my friend
Here is the error I got.
Error in loadNamespace(i, c(lib.loc, .libPaths())) :
there is no package called ‘munsell’
I found no munsell package in CRAN.
Hi Julie,
That’s strange. This isn’t a required package for the plot, but check it out anyway:
http://cran.r-project.org/web/packages/munsell/index.html
-Marcus
Hi, thanks for your post! I try to use it in classification model but it’s failed. Does it only for regression? Thanks!
Hi,
The function is meant for neural network models created using the nnet package. It will not work for other model types.
-Marcus
Visualizing neural networks from the nnet package – R is my friend | luiz p. c. de freitas
Variable importance in neural networks – R is my friend
Hi, your graph is great! How to use this plot function for a neural network without a hidden layer? doenst seems to work.
Error: Error in matrix(seq(1, layers[1] * layers[2] + layers[2]), ncol = layers[2]) :
data is too long
Thanks! The function only works with model objects from the nnet package. As far as I know, the package cannot create neural networks w/o a hidden layer.
Congratulations for the function. I got the same error.
Actually when you put size = 0 and skip = true it will be a neural network without hidden layer. By the way, on my experiments this configuration reach the best results comparing with nnets with hidden layer.
Hi Andre,
Thanks, I didn’t know you could do that with the nnet package. Interesting comment though about your model – I think in lots of cases a simple model is sufficient or better than a neural network model. Just because a model seems fancy (e.g., a neural network) doesn’t mean it’s the best approach for a particular dataset!
-Marcus
Hi in your plot, for example if the weight is 0, but there is still a line plotted. Is there anyway to change this?
Very nice write up, thank you for doing this.
Are you planning to adopt it for RSNNS as well?
Thanks, probably not since I haven’t done any work with that package. However, the implementation may be straightforward if there’s a convenient way to get the weights.
Would you be available for some questions about your code perhaps? Think I will try to modify it for RSNNS.
Thanks for the helpful blog! I enjoyed learning it. I’m new to R but I have been working on NNs in python. In your code you can extract the weights of your plot. I wonder it I can do this the other way around. I have matrices of weights from large NN python simulations that i want to visualize without re-running the NN in R.
That would be possible by changing the code for the plotting function. However, this may be difficult since you’re new to R. The problem is that the variable names, weights, and nnet structure are all extracted from the input to the function, which is a model object from nnet. Inputting the weights directly would mess up the workflow for the function. I’ve been thinking about improving the flexibility of the function, so I will consider this request for future changes. Stay tuned!
Yeah, the syntax is just so different from other languages, so I’m studying the basics of R now including neuralnet for visualization. I’m looking forward to the future modifications of the function!
Visualizing neural networks in R – update – R is my friend
Thanks for your codes, but I have some categorical predictors, how could I show the class of these predictors? Also, the output of my model doesn’t show the label.
I use this code
model = nnet(y ~ ., data = trainingset,size = 3,maxit = 100)
Thanks
Hi Vanessa, have you tried the updated plotting function here? I modified it a while back to correctly plot categorical variables. You can also specify the label names with the
x.labandy.labarguments.Yeah, I got what I want! Thank you so much!
In mod2 (using neuralnet package) I get the following warning message.
Warning message: algorithm did not converge in 1 of 1 repetition(s) within the stepmax
Can you please help with this?
Thanks
Hi there, I was wondering when someone would notice that… I’ve had some trouble fitting models with continuous response variable with the neuralnet package. The default model fitting function is not the back-propagation algorithm, which I’m sure has to do with the convergence issue. I tried setting the
algorithmargument to ‘backprop’. This also requires a value for thelearningrateargument. I tried a few different values, but was never able to get the model to work.Anyhow, try the function with the example from the neuralnet package:
library(devtools) library(neuralnet) source_url('https://gist.github.com/fawda123/7471137/raw/c720af2cea5f312717f020a09946800d55b8f45b/nnet_plot_update.r') AND <- c(rep(0,7),1) OR <- c(0,rep(1,7)) binary.data <- data.frame(expand.grid(c(0,1), c(0,1), c(0,1)), AND, OR) print(net <- neuralnet(AND+OR~Var1+Var2+Var3, binary.data, hidden=0, rep=10, err.fct="ce", linear.output=FALSE)) plot.nnet(net)Thanks a lot for the quick reply. This piece of code works fine.
I have another question(not related to your post though). How to change the functional form to calculate the weights and output variables? According to my knowledge, neural network tries to fit in a linear regression model when supplied with continuous input and output variables. I’m looking for a way to change the functional form, so that it fits a nonlinear model.
Neural networks are basically large non-linear regression models in the sense that the model is describing relationships between predictor and response variables using multiple parameters/weights. I’ve read that a network with no hidden layer is analogous to a linear regression model, i.e., one weight defines the link between each input node to the output like a slope parameter in a regression. Any time you fit a neural network with at least one or more nodes in the hidden layer, you are fitting a non-linear model. If you’re talking about the activation function, that can usually be changed with one of the arguments in the model function, e.g.
linOutargument formlp. This function is applied to the output for each neuron/node (i.e., hidden or output neuron) as a sort of transformation. In my experience, the activation function is usually chosen as a linear or logistic/sigmoidal output. To be honest, I’m not entirely certain how this affects the model precision but I know it depends on the form of your response variable, i.e., categorical output or continuous values on the range of 0–1 should use logistic activation functions, all others should be continuous. Note that using a linear activation function still results in a ‘non-linear model’ if a hidden layer with multiple nodes is used, i.e., the linear output from each node interacts in non-linear ways to influence the predicted response.Terrific! This should definitely get into the core “nnet” package man!
Thanks! I’m working on developing a package for the plot, sensitivity, and variable importance functions. Stay tuned!
i dont find mod.in in my nnet package. Between, is sensitivity analysis work for time series data? its interesting to know the correlation input and the response. tq!
Hello,
The plot function should work with models created with nnet. The examples above show how this can be done. For time series data, the sensitivity analysis should work but I can’t say for sure. I don’t have much experience using neural networks with time series.
-Marcus
Hello, i am currently doing forecasting using neural network. Is there anyway that i can update every forecaste value and use it to predict the future value??
Hi Tiger,
I don’t have much experience using neural networks with time series – maybe have a look here for resources.
-Marcus
This is great! The link to the code snippet is stale now that github has gists. New link is https://gist.githubusercontent.com/fawda123/5086859/raw/cc1544804d5027d82b70e74b83b3941cd2184354/nnet_plot_fun.r
Thanks for pointing that out, I’ve changed the url above.
Your code was very useful. I initially tried the code from another page, http://www.r-bloggers.com/visualizing-neural-networks-from-the-nnet-package/
The code from above page threw an error. It showed NULL for ‘eval(parse(text = script))’.
The code from this page worked fine. I guess its due to the different URL used here.
Hi Vivek,
Take a look at the NeuralNetTools package on CRAN: https://cran.r-project.org/web/packages/NeuralNetTools/index.html
It will have a stable, working version of the plotting function described in the blog.
-Marcus
Hi
Your Code helped me alot. Thank you so much.
Just 2 doubts:
1. In gar.fun: is there a way to plot importance of some definite variables like 2 or 3. Especially required for Big Data
2. In plot.nnet: you showed how to plot a single connection from input to output variable. In “all.in” I entered a vector of 6 input variables, but it plotted only 2 and also if there can be a way to remove all other non required variables. Just plot these 6.
Hi Ishan, thanks for checking out my blog. Have you tried using the package released on CRAN? That might help with some of your problems.
https://cran.r-project.org/web/packages/NeuralNetTools/index.html
-Marcus
HI Marcus.
Thanks for the reply
Actually I have tried NNTools: garson,olden, plotnnet and lekprofile.
Garson: Only the rel imp in form of positive values is generated (but that was solved in gar.fun).
In garson, gar.fun, olden: I am still not able to select particular input variables for plot. Because of which I have to keep the argument ‘bar.plot=F’ to get a tabular version for further analysis.
Only in lekprofile I am able to select the particular variable for sensitivity
Hi, how do you (typically) select a representative network to plot given many thousands ANNs were built for each response variable?
Thanks
jO
Hi jO, that depends on your goal of selecting the representative network. Are you creating thousands of networks to identify the “best” model or are you simply creating thousands for model averaging? If the former, then it should be obvious which network to choose. If the latter, it may not be appropriate to show a representative network because one may not exist. Consider showing a few as examples to demonstrate that you are using a multi-model approach for each response variable. It might not matter which model you show because the emphasis is on the predictive power of the combined models. Does that make sense?
Hi Marcus, thanks for your reply. I’m reading a paper and I think they’re doing the former. They built thousands of ANNs for each response variable and were scored based on Pearson correlation coeffs. They selected a single best ANN for each response based on the single highest correlation coeff. Wouldn’t it be better to treat the correlation coeffs as a distribution and choose some mode of this distribution as the “best” or more likely ANN?
Yea I would be cautious in picking the “best” solely from a single correlation coefficient or any other performance measure. The performance of neural networks is highly sensitive to the training data and initial conditions (e.g., random starting weights), so I would imagine that the highest ranking model could give similar results as any of a number of the other models. I agree that looking for commonalities among several potential candidate models would be better, as in your suggested approach.
Hi, i am try to use the code but whe i run failed
script<-getURL(raw.fun, ssl.verifypeer = FALSE)
Error in function (type, msg, asError = TRUE) :
error:1407742E:SSL routines:SSL23_GET_SERVER_HELLO:tlsv1 alert protocol version
In addition, we can fine the funtion plot.nnet, we use de many libraries (NeuralNetTools, gamlss.add, and nnet) but not run. Cuuld you help me?
Hi Joaquin, please use the functions in the NeuralNetTools package. They are more current than the functions in my blog. Check out the publication for more info: https://www.jstatsoft.org/article/view/v085i11